Cellular and viral determinants of the hepatitis C virus replication compartment
Principal investigator: Bartenschlager
One hallmark of positive-strand RNA viruses is the extensive reorganization of intracellular membrane compartments. In the case of the hepatitis C virus (HCV), ER membranes are remodeled to create a replication organelle (RO) that is composed predominantly of double membrane vesicles (DMVs) residing in close proximity of lipid droplets (LDs). The molecular details how these membranes are reorganized and how they serve the purpose of viral RNA replication and assembly of infectious virus particles are poorly understood. It is the aim of this project to dissect the viral and cellular determinants involved in the biogenesis of the HCV RO, its link to LDs and how these structures and organelles contribute to the assembly of virions. To this end, four research lines will be followed:
- Based on lipidome and proteome analyses of ROs isolated from HCV-replicating cells we performed a guided siRNA screen which revealed several hits with a strong phenotype on viral RNA replication or assembly. We aim to investigate the role of these proteins in RO formation and RO (replicase) activity at the molecular level.
- In accordance with previous results by us and others we want to study the mechanism how HCV utilizes autophagy components for its replication cycle and identify the very early steps of HCV RO formation by employing CLEM.
- Since the HCV protein NS4B is critically involved in RO formation, we aim to study the role by which a possible NS4B viroporin activity contributes to RO biogenesis, HCV replicase activity and possibly the assembly of infectious HCV particles.
- LDs are essential for HCV assembly and envelopment of virus particles is coordinated by the HCV protein NS2 that binds both the viral replicase containing the RNA genome and the envelope glycoproteins. Moreover, by using cholesterol binding experiments, we obtained preliminary evidence that NS2 strongly binds to this lipid. Therefore, we will map the cholesterol binding domain in NS2, conduct reverse and forward genetics screens of NS2 for cholesterol interaction and HCV assembly and establish a NS2-based live cell imaging system to monitor the assembly. Finally, we want to use this set-up to study how apolipoprotein E, an integral component of infectious HCV particles, gets incorporated into virions and which role this apolipoprotein plays for virus assembly and egress.
Figure legend:
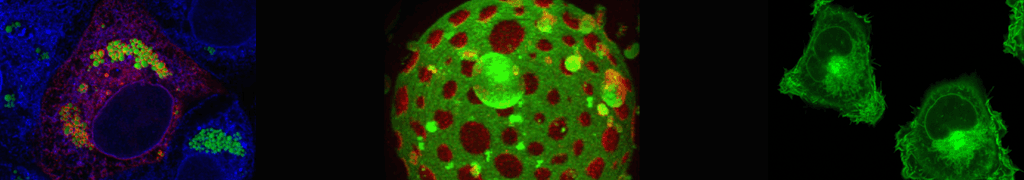
Model of HCV replication and assembly. Viral RNA is amplified in double membrane vesicles (DMV) and delivered, probably with the involvement of viral proteins, to sites of virion assembly. There particles form by budding into the ER lumen. On the surface of lipid droplets, the HCV proteins NS5A and core accumulate and they might be involved in the assembly of infectious virus particles. Budding into the ER lumen is thought to be promoted by the envelope glycoproteins E1 and E2 in conjunction with p7 and NS2. It is assumed that already during assembly HCV particles associate with lipids and apolipoproteins (not shown). Figure is modified from Zayas et al., Plos Pathogens, 2016.